Missing Data
CRD 150 - Quantitative Methods in Community Research
Professor Noli Brazil
This guide outlines what to do when you encounter missing values in your data set. This is not meant to be a comprehensive treatment of how to deal with missing data. One can teach a whole class or write a whole book on this subject. Instead, the guide provides a brief overview, complementing Handout 2.
Bringing data into R
First, let’s load in the required packages for this guide. Depending on when you read this guide, you may need to install some of these packages using the function install.packages() before calling library(). You may also encounter some unfamiliar packages. In particular, we are using spatial data in this lab, and hence using packages that deal with the processing and analysis of spatial data.
library(sf)
library(sp)
library(spdep)
library(tidyverse)
library(tmap)If you are reading this guide before the class has not gone through the introduction to spatial data and spatial clustering labs, don’t pay too much attention to the why and how certain spatial functions work, and instead focus on the methods and functions that are pertinent to dealing with missing data in R. These functions can be used on both spatial and nonspatial data.
We’ll be using the shapefile saccity.shp. The file contains Sacramento City census tracts with the percent of the tract population living in subsidized housing, which was taken from the U.S. Department of Housing and Urban Development data portal.
We’ll need to read in the shapefile. First, set your working directory to a folder you want to save your data in.
setwd("path to the folder containing saccity.shp")I saved the file in Github as a zip file. Download that zip file using the function download.file(), unzip it using the function unzip(), and read the file into R using st_read()
download.file(url = "https://raw.githubusercontent.com/crd150/data/master/saccity.zip", destfile = "saccity.zip")
unzip(zipfile = "saccity.zip")
sac.city.tracts.sf <- st_read("saccity.shp")## Reading layer `saccity' from data source `/Users/noli/Documents/UCD/teaching/CRD150/Lab/crd150.github.io/saccity.shp' using driver `ESRI Shapefile'
## Simple feature collection with 120 features and 3 fields
## geometry type: MULTIPOLYGON
## dimension: XY
## bbox: xmin: -121.5601 ymin: 38.43757 xmax: -121.3627 ymax: 38.6856
## epsg (SRID): 4269
## proj4string: +proj=longlat +ellps=GRS80 +towgs84=0,0,0,0,0,0,0 +no_defsIn case you are having problems with the above code, try installing the package utils. If you are still having problems, download the zip file from Canvas (Additional data). Save that file into the folder you set your working directory to. Then use st_read().
Let’s look at the tibble.
sac.city.tracts.sf## Simple feature collection with 120 features and 3 fields
## geometry type: MULTIPOLYGON
## dimension: XY
## bbox: xmin: -121.5601 ymin: 38.43757 xmax: -121.3627 ymax: 38.6856
## epsg (SRID): 4269
## proj4string: +proj=longlat +ellps=GRS80 +towgs84=0,0,0,0,0,0,0 +no_defs
## First 10 features:
## GEOID NAME psubhous
## 1 06067000100 Census Tract 1, Sacramento County, California NA
## 2 06067000200 Census Tract 2, Sacramento County, California NA
## 3 06067000300 Census Tract 3, Sacramento County, California NA
## 4 06067000400 Census Tract 4, Sacramento County, California 2.18
## 5 06067000500 Census Tract 5, Sacramento County, California 7.89
## 6 06067000600 Census Tract 6, Sacramento County, California 15.44
## 7 06067000700 Census Tract 7, Sacramento County, California 17.00
## 8 06067000800 Census Tract 8, Sacramento County, California 9.17
## 9 06067001101 Census Tract 11.01, Sacramento County, California 5.24
## 10 06067001200 Census Tract 12, Sacramento County, California 10.05
## geometry
## 1 MULTIPOLYGON (((-121.4472 3...
## 2 MULTIPOLYGON (((-121.4505 3...
## 3 MULTIPOLYGON (((-121.4615 3...
## 4 MULTIPOLYGON (((-121.4748 3...
## 5 MULTIPOLYGON (((-121.487 38...
## 6 MULTIPOLYGON (((-121.487 38...
## 7 MULTIPOLYGON (((-121.5063 3...
## 8 MULTIPOLYGON (((-121.5116 3...
## 9 MULTIPOLYGON (((-121.4989 3...
## 10 MULTIPOLYGON (((-121.48 38....And make sure it looks like Sacramento city
tm_shape(sac.city.tracts.sf) +
tm_polygons()
Cool? Cool.
Summarizing data with missing values
The variable psubhous gives us the percent of the tract population living in subsidized housing. What is the mean of this variable?
sac.city.tracts.sf %>% summarize(mean = mean(psubhous))## Simple feature collection with 1 feature and 1 field
## geometry type: POLYGON
## dimension: XY
## bbox: xmin: -121.5601 ymin: 38.43757 xmax: -121.3627 ymax: 38.6856
## epsg (SRID): 4269
## proj4string: +proj=longlat +ellps=GRS80 +towgs84=0,0,0,0,0,0,0 +no_defs
## mean geometry
## 1 NA POLYGON ((-121.409 38.50368...Oh oh, no bueno. We get “NA”. If a variable has an NA, most R functions summarizing that variable will automatically yield an NA. This means we have missing data.
What if we try to do some kind of spatial data analysis on the data? Surely, R won’t give us a problem, right? Let’s calculate the Moran’s I for psubhous.
#Turn sac.city.tracts.sf into an sp object.
sac.city.tracts.sp <- as(sac.city.tracts.sf, "Spatial")
sacb<-poly2nb(sac.city.tracts.sp, queen=T)
sacw<-nb2listw(sacb, style="W")
moran.test(sac.city.tracts.sp$psubhous, sacw) ## Error in na.fail.default(x): missing values in objectSimilar to nonspatial data functions, R will force you to deal with your missing data values. In this case, R gives us an error.
Summarizing the extent of missingness
Before you do any analysis on your data, it’s a good idea to check the extent of missingness in your data set. The best way to do this is to use the function aggr(), which is a part of the VIM package. Install this package if you have not already done so.
install.packages("VIM")Load it into your current R session.
library(VIM)Then run the aggr() function as follows
summary(aggr(sac.city.tracts.sf))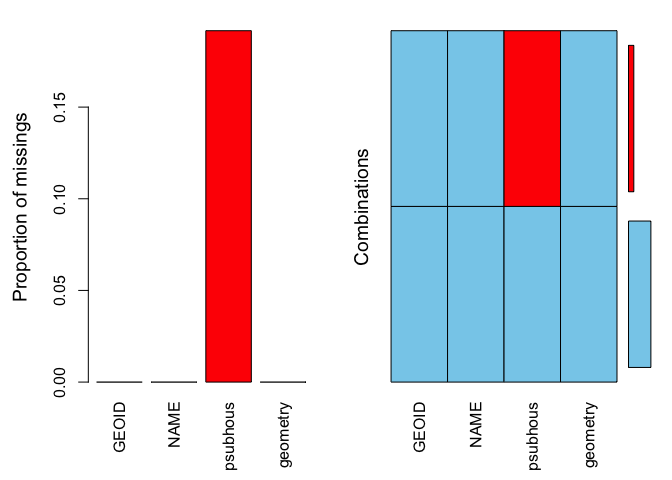
##
## Missings per variable:
## Variable Count
## GEOID 0
## NAME 0
## psubhous 23
## geometry 0
##
## Missings in combinations of variables:
## Combinations Count Percent
## 0:0:0:0 97 80.83333
## 0:0:1:0 23 19.16667The results show two tables and two plots. The left-hand side plot shows the proportion of cases that are missing values for each variable in the data set. The right-hand side plot shows which combinations of variables are missing. The first table shows the number of cases that are missing values for each variable in the data set. The second table shows the percent of cases missing values based on combinations of variables. The results show that 23 or 19% of census tracts are missing values on the variable psubhous.
Exclude missing data
Recall from Handout 2 that the two basic methods for dealing with missing values are: (1) Ignore the observations with missing values or (2) Impute the mean.
You can implement method (1) two ways. In the first way, you physically remove the offending missing value tracts from the data set. You do this using the drop_na() command.
sac.city.tracts.sf.rm <- drop_na(sac.city.tracts.sf, psubhous)You now get your mean
sac.city.tracts.sf.rm %>% summarize(mean = mean(psubhous))## Simple feature collection with 1 feature and 1 field
## geometry type: POLYGON
## dimension: XY
## bbox: xmin: -121.5601 ymin: 38.43757 xmax: -121.3627 ymax: 38.68522
## epsg (SRID): 4269
## proj4string: +proj=longlat +ellps=GRS80 +towgs84=0,0,0,0,0,0,0 +no_defs
## mean geometry
## 1 6.13866 POLYGON ((-121.409 38.50368...And your Moran’s I
#Turn sac.city.tracts.sf into an sp object.
sac.city.tracts.sp.rm <- as(sac.city.tracts.sf.rm, "Spatial")
sacb.rm<-poly2nb(sac.city.tracts.sp.rm, queen=T)
sacw.rm<-nb2listw(sacb.rm, style="W")
moran.test(sac.city.tracts.sp.rm$psubhous, sacw.rm) ##
## Moran I test under randomisation
##
## data: sac.city.tracts.sp.rm$psubhous
## weights: sacw.rm
##
## Moran I statistic standard deviate = 1.5787, p-value = 0.0572
## alternative hypothesis: greater
## sample estimates:
## Moran I statistic Expectation Variance
## 0.079875688 -0.010416667 0.003271021It’s often a better idea to keep your data intact rather than remove cases. For many of R’s functions, there is an na.rm = TRUE option, which tells R to remove all cases with missing values on the variable when performing the function. For example, inserting the na.rm = TRUE option in the mean() function yields
sac.city.tracts.sf %>% summarize(mean = mean(psubhous, na.rm=TRUE))## Simple feature collection with 1 feature and 1 field
## geometry type: POLYGON
## dimension: XY
## bbox: xmin: -121.5601 ymin: 38.43757 xmax: -121.3627 ymax: 38.6856
## epsg (SRID): 4269
## proj4string: +proj=longlat +ellps=GRS80 +towgs84=0,0,0,0,0,0,0 +no_defs
## mean geometry
## 1 6.13866 POLYGON ((-121.409 38.50368...In the function moran.test(), we use the option na.action=na.omit
moran.test(sac.city.tracts.sp$psubhous, sacw, na.action=na.omit) ##
## Moran I test under randomisation
##
## data: sac.city.tracts.sp$psubhous
## weights: sacw
## omitted: 1, 2, 3, 17, 19, 21, 22, 23, 34, 43, 44, 46, 49, 50, 69, 74, 75, 99, 100, 101, 102, 106, 120
##
## Moran I statistic standard deviate = 1.5787, p-value = 0.0572
## alternative hypothesis: greater
## sample estimates:
## Moran I statistic Expectation Variance
## 0.079875688 -0.010416667 0.003271021Impute the mean
Usually, it is better to keep observations than discard or ignore them, so strategy (2), imputing the mean, is often the better approach. In the case of psubhous, were missing almost 20% of the data, which is a lot of cases to exclude. Moreover, not all functions have the built in na.rm or na.action options. Also, look at the map of the sf object without the tracts missing data.
tm_shape(sac.city.tracts.sf.rm) + tm_polygons(col="blue")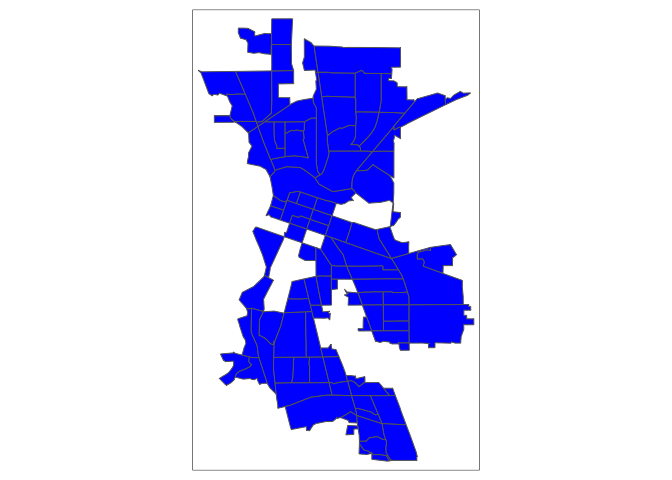
We’ve got permanent holes in Sacramento because we’ve physically removed census tracts with missing values. Of course, we can shade the missing tracts a certain color and label this in the legend. We’ll do that in Lab 4. But, let’s see how imputing the mean works.
To impute the mean, we need to use the tidy friendly impute_mean() function in the tidyimpute package. Install this package if you haven’t already done so.
install.packages("tidyimpute")and load it into R.
library(tidyimpute)Then use the function impute_mean(). To use this function, pipe in the data set and then type in the variables you want to impute.
sac.city.tracts.sf.mn <- sac.city.tracts.sf %>%
impute_mean(psubhous)Note that you can impute more than one variable within impute_mean() using the comma separator.
We should now have no missing values
summary(aggr(sac.city.tracts.sf.mn))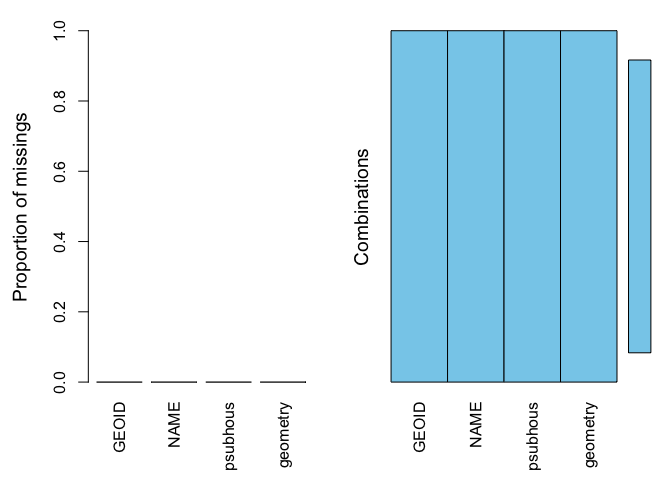
##
## Missings per variable:
## Variable Count
## GEOID 0
## NAME 0
## psubhous 0
## geometry 0
##
## Missings in combinations of variables:
## Combinations Count Percent
## 0:0:0:0 120 100Therefore allowing us to calculate the mean of psubhous
sac.city.tracts.sf.mn %>% summarize(mean = mean(psubhous))## Simple feature collection with 1 feature and 1 field
## geometry type: POLYGON
## dimension: XY
## bbox: xmin: -121.5601 ymin: 38.43757 xmax: -121.3627 ymax: 38.6856
## epsg (SRID): 4269
## proj4string: +proj=longlat +ellps=GRS80 +towgs84=0,0,0,0,0,0,0 +no_defs
## mean geometry
## 1 6.13866 POLYGON ((-121.409 38.50368...And your Moran’s I
#Turn sac.city.tracts.sf into an sp object.
sac.city.tracts.sp.mn <- as(sac.city.tracts.sf.mn, "Spatial")
sacb.mn<-poly2nb(sac.city.tracts.sp.mn, queen=T)
sacw.mn<-nb2listw(sacb.mn, style="W")
moran.test(sac.city.tracts.sp.mn$psubhous, sacw.mn) ##
## Moran I test under randomisation
##
## data: sac.city.tracts.sp.mn$psubhous
## weights: sacw.mn
##
## Moran I statistic standard deviate = 1.8814, p-value = 0.02996
## alternative hypothesis: greater
## sample estimates:
## Moran I statistic Expectation Variance
## 0.081778772 -0.008403361 0.002297679And our map has no holes!!
tmap_mode("view")
tm_shape(sac.city.tracts.sf.mn) +
tm_polygons(col = "psubhous", style = "quantile")Other Imputation Methods
The tidyimpute package has a set of functions for imputing missing values in your data. The functions are categorized as univariate and multivariate, where the former imputes a single value for all missing observations (like the mean) whereas the latter imputes a value based on a set of non-missing characteristics. Univariate methods include
impute_max- maximumimpute_minimum- minimumimpute_median- median valueimpute_quantile- quantile valueimpute_sample- randomly sampled value via bootstrap
Multivariate methods include
impute_fit,impute_predict- use a regression model to predict the valueimpute_by_group- use by-group imputation
Some of the multivariate functions may not be fully developed at the moment, but their test versions may be available for download. These methods go beyond the scope of this class, but you can check a number of user created vignettes, include the official R vignette.

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Website created and maintained by Noli Brazil